Check out the latest publication from the Mehl/Cooley & Barbar Labs!
Accessing isotopically labeled proteins containing genetically encoded phosphoserine for NMR with optimized expression conditions
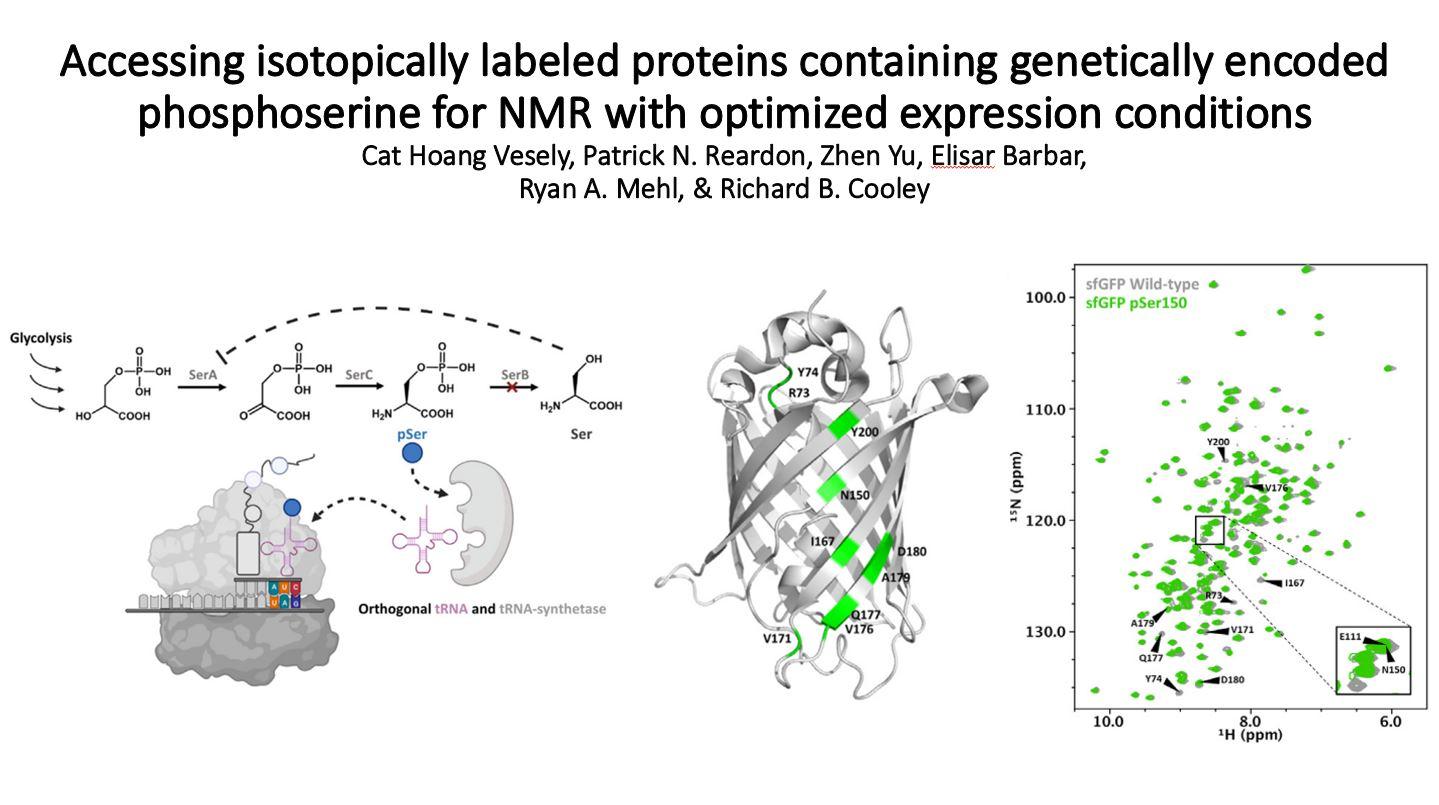
Phosphoserine (pSer) sites are primarily located within disordered protein regions, making it difficult to experimentally ascertain their effects on protein structure and function. Therefore, the production of 15N- (and 13C)-labeled proteins with site-specifically encoded pSer for NMR studies is essential to uncover molecular mechanisms of protein regulation by phosphorylation. While genetic code expansion technologies for the translational installation of pSer in Escherichia coli are well established and offer a powerful strategy to produce site specifically phosphorylated proteins, methodologies to adapt them to minimal or isotope-enriched media have not been described. This shortcoming exists because pSer genetic code expansion expression hosts require the genomic ΔserB mutation, which increases pSer bioavailability but also imposes serine auxotrophy, preventing growth in minimal media used for isotopic labeling of recombinant proteins. Here, by testing different media supplements, we restored normal BL21(DE3) ΔserB growth in labeling media but subsequently observed an increase of phosphatase activity and mis-incorporation not typically seen in standard rich media. After rounds of optimization and adaption of a high-density culture protocol, we were able to obtain ≥10 mg/L homogenously labeled, phosphorylated superfolder GFP. We propose this cost-effective methodology will pave the way for more routine access to pSer-enriched proteins for 2D and 3D NMR analyses.