Check out the latest publication from the Freitag group!
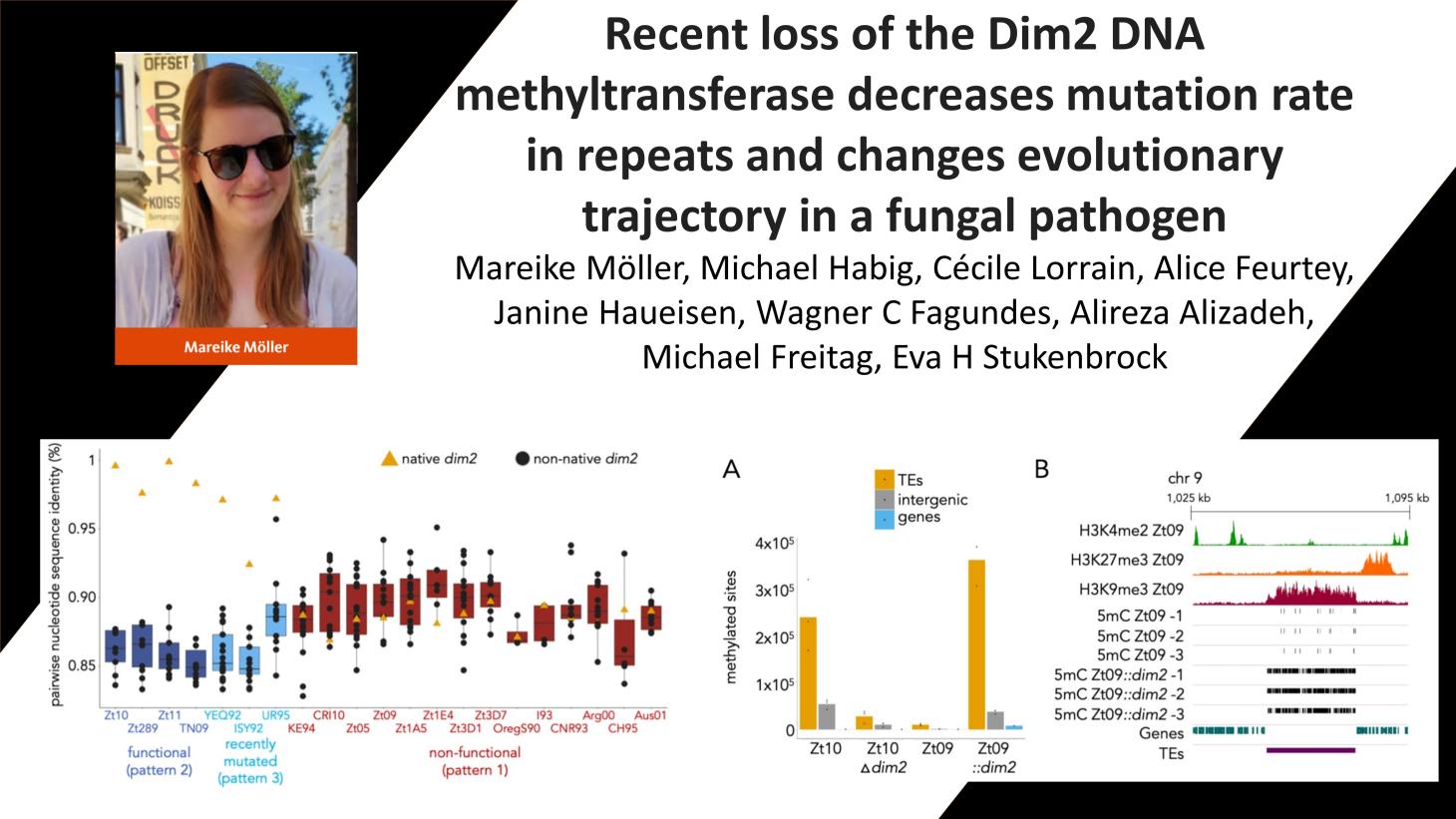
Cytosine DNA methylation (5mC) is known to silence transposable elements in fungi and thereby appears to contribute to genome stability. The genomes of plant pathogenic fungi are highly diverse, differing substantially in transposon content and distribution. Here, we show extensive differences of 5mC levels within a single species of an important wheat pathogen. These differences were caused by inactivation of the DNA methyltransferase Dim2 in the majority of studied isolates. Presence of widespread 5mC increased point mutation rates in regions with active or mutated transposable elements during mitosis. The mutation pattern is dependent on the presence of Dim2 and resembles a mitotic version of Repeat-Induced Point mutation (RIP). Thus, loss of 5mC may represent an evolutionary trade-off offering adaptive potential at the cost of transposon control.